Whole genome sequencing
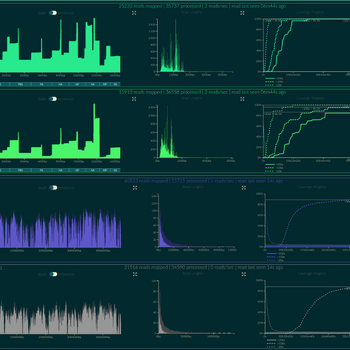
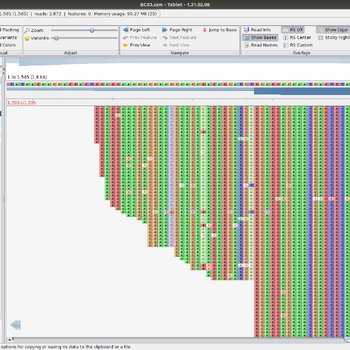
We offer whole genome sequencing of bacteria, which is performed by the Molecular Biology Department using the new generation Oxford Nanopore method. This method is one of the most advanced third-generation sequencing technologies.
The method is based on the principle that a nucleic acid passes through a bacterial pore and thereby produces a change in electrical signal depending on the nucleotide sequence. The analysis takes place on a chip (called a flow cell) that contains a lipid membrane with anchored nanopores. For more information on this technology, visit the Oxford Nanopore website.
A major benefit of sequencing using Oxford Nanopore technology is the ability to read very long segments of DNA/RNA. The length of the segment read depends only on the length of the nucleic acid obtained.
Other advantages of this sequencing are:
speed of library preparation
Some kits do not use PCR amplification and therefore do not have the errors associated with PCR.
the accuracy of the output data
Due to advances in bioinformatics, chemistry and the flow cell R generation offered, accuracy is reported to be >99%, which is on par with the accuracy of Illumina sequencing data.
low requirements for number of samples per run
E.g. compared to Illumina sequencing
We have applied this sequencing technology for whole genome sequencing of SARS_Cov2 and are currently using it for whole genome sequencing of avian influenza virus.
We are now offering the method for sequencing bacterial genomes as raw data.
For the bacterial species listed below, we also offer sequence assembly (bacterial chromosome and plasmids). Serotyping and MLST profiling and analysis of genes for toxins, resistances, etc. can be performed by appointment.
- Escherichia coli (EC)
- Salmonella
- Clostridium perfringens
- Taylorella equigenitalis
- Enterococcus faecalis
- Enterococcus faecium
- Campylobacter jejuni
- Staphylococcus aureus